Interpretation Expert Settings¶
The following is a list of the Interpretation expert settings that are available when setting up a new interpretation from the MLI page. The name of each setting is preceded by its config.toml label.
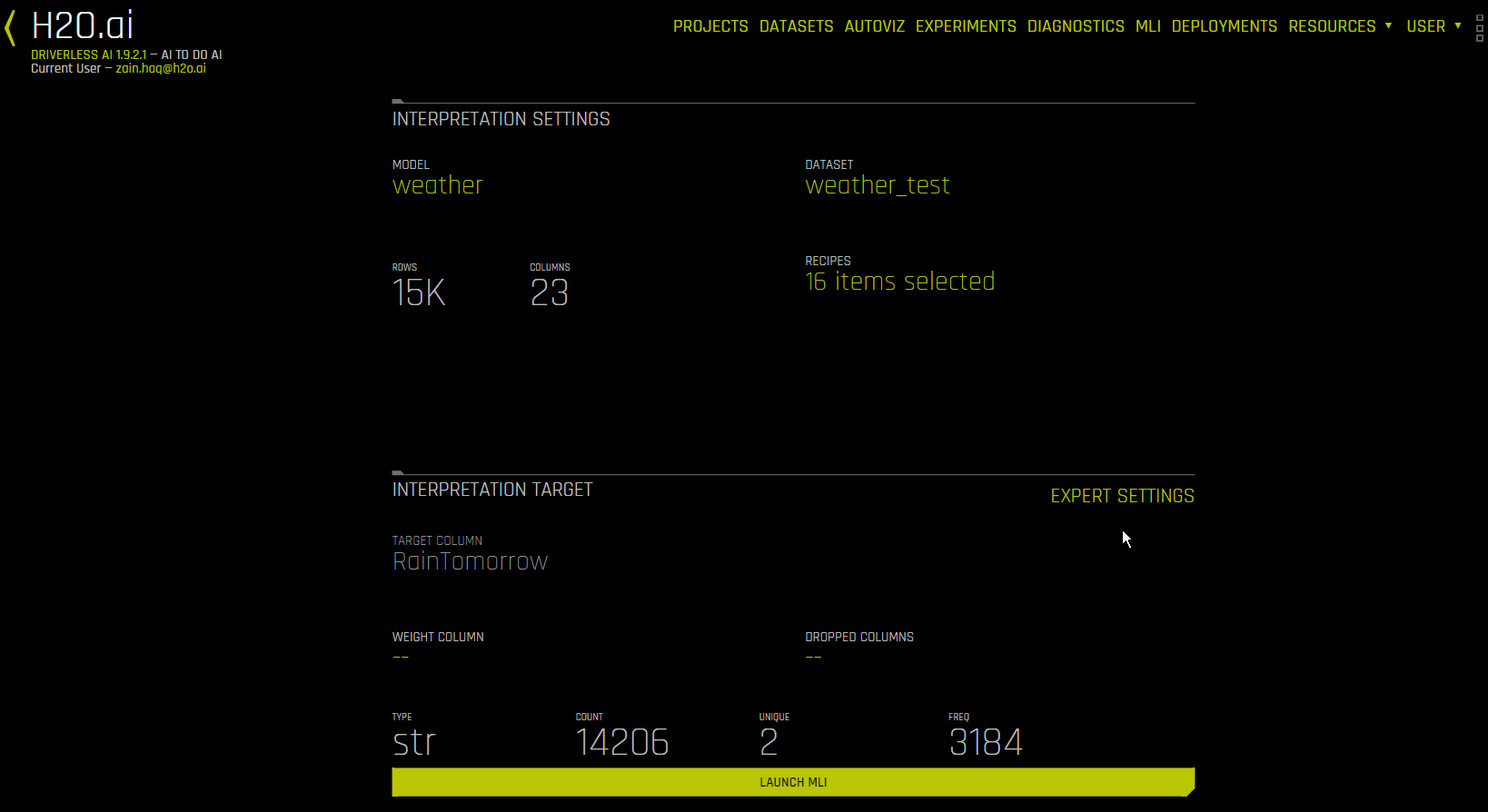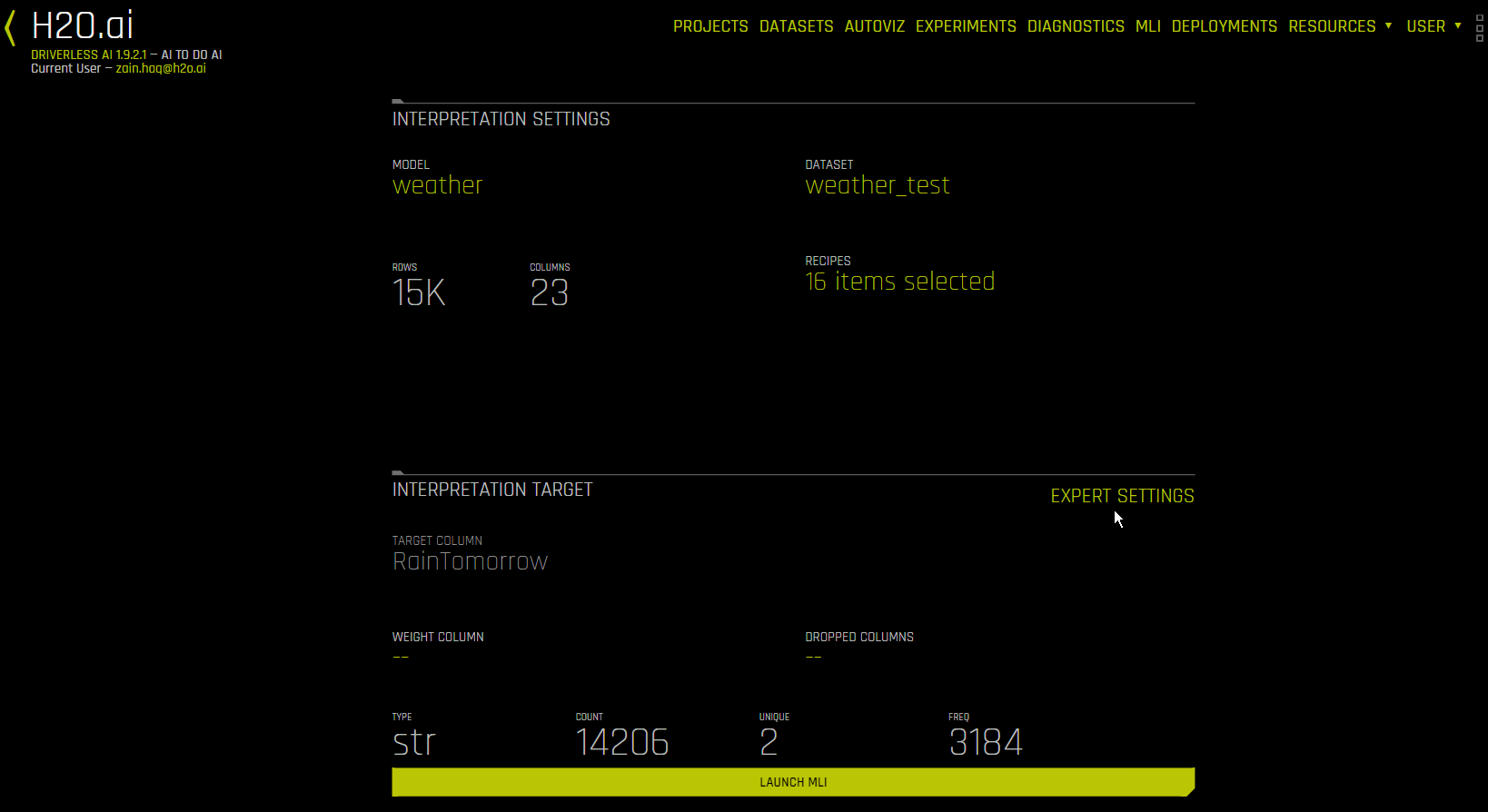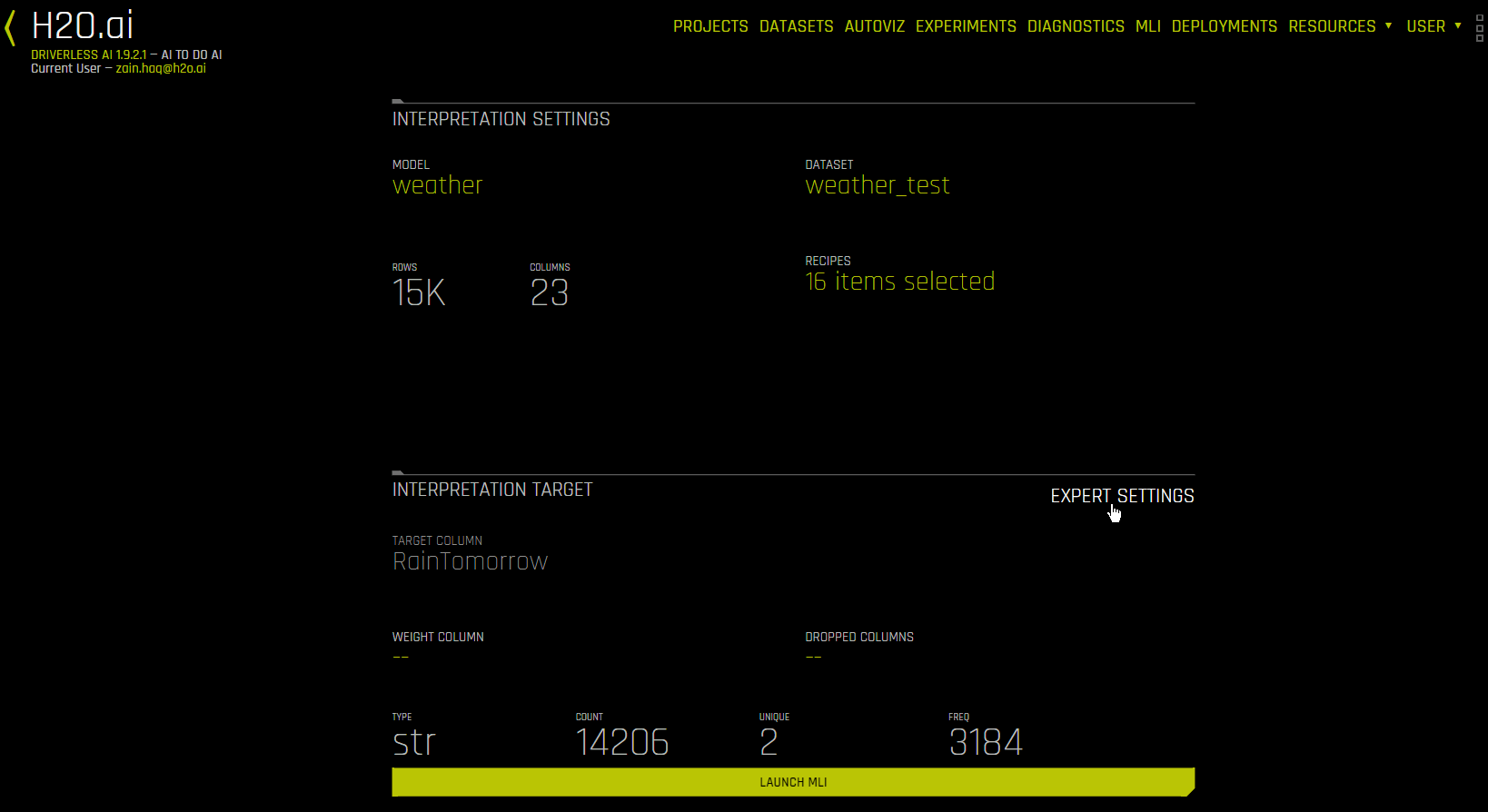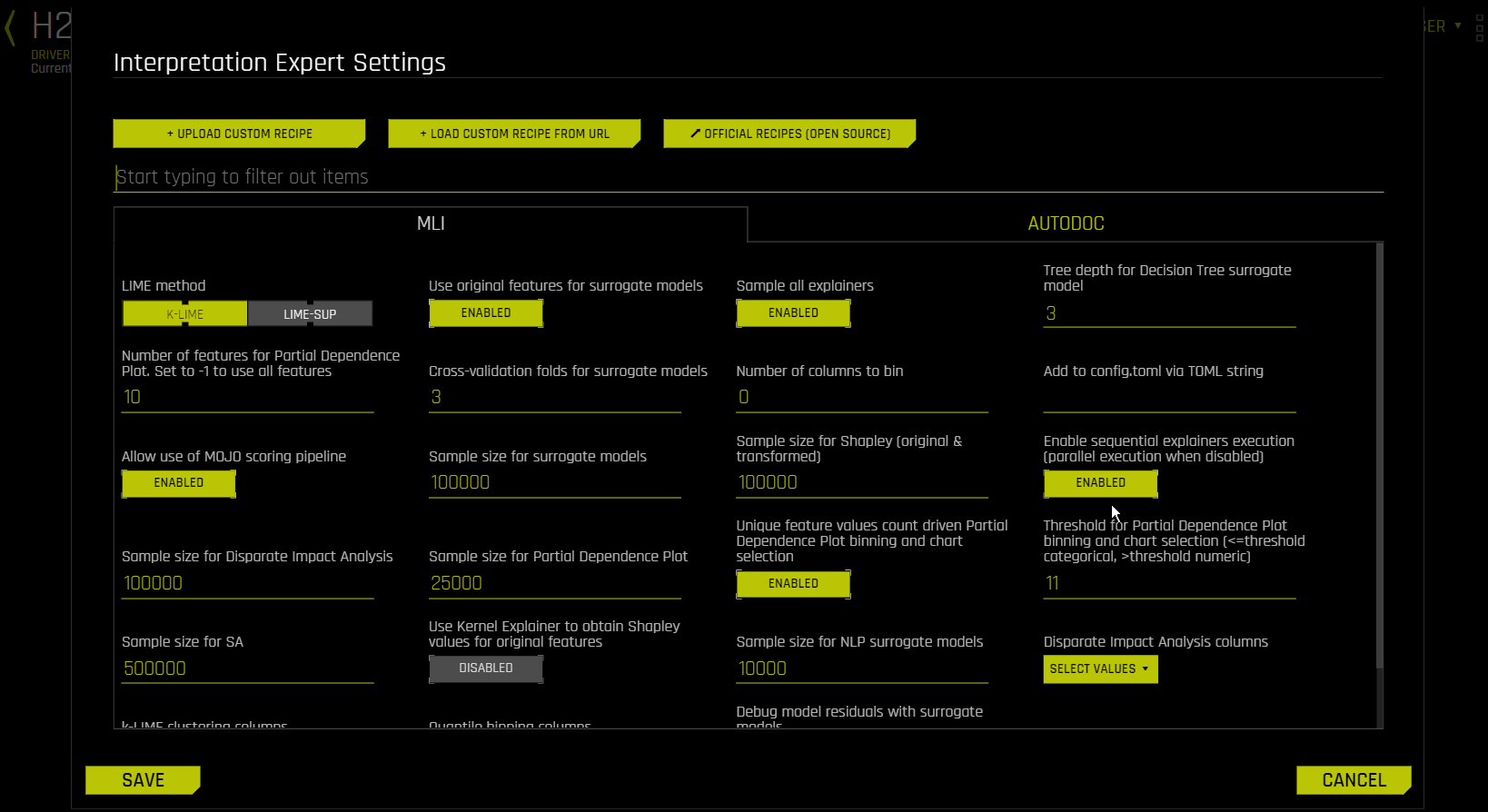
MLI Tab¶
mli_lime_method¶
LIME Method
Select a LIME method of either K-LIME (default) or LIME-SUP.
K-LIME (default): creates one global surrogate GLM on the entire training data and also creates numerous local surrogate GLMs on samples formed from k-means clusters in the training data. The features used for k-means are selected from the Random Forest surrogate model’s variable importance. The number of features used for k-means is the minimum of the top 25% of variables from the Random Forest surrogate model’s variable importance and the max number of variables that can be used for k-means, which is set by the user in the config.toml setting for
mli_max_number_cluster_vars. (Note, if the number of features in the dataset are less than or equal to 6, then all features are used for k-means clustering.) The previous setting can be turned off to use all features for k-means by settinguse_all_columns_klime_kmeansin the config.toml file totrue. All penalized GLM surrogates are trained to model the predictions of the Driverless AI model. The number of clusters for local explanations is chosen by a grid search in which the \(R2\) between the Driverless AI model predictions and all of the local K-LIME model predictions is maximized. The global and local linear model’s intercepts, coefficients, \(R2\) values, accuracy, and predictions can all be used to debug and develop explanations for the Driverless AI model’s behavior.LIME-SUP: explains local regions of the trained Driverless AI model in terms of the original variables. Local regions are defined by each leaf node path of the decision tree surrogate model instead of simulated, perturbed observation samples - as in the original LIME. For each local region, a local GLM model is trained on the original inputs and the predictions of the Driverless AI model. Then the parameters of this local GLM can be used to generate approximate, local explanations of the Driverless AI model.
mli_use_raw_features¶
Use Original Features for Surrogate Models
Specify whether to use original features or transformed features in the surrogate model for the new interpretation. This is enabled by default.
Note: When this setting is disabled, the K-LIME clustering column and quantile binning options are unavailable.
mli_sample¶
Sample All Explainers
Specify whether to perform the interpretation on a sample of the training data. By default, MLI will sample the training dataset if it is greater than 100k rows. (The equivalent config.toml setting is mli_sample_size.) This is enabled by default. Turn this toggle off to run MLI on the entire dataset.
mli_dt_tree_depth¶
Tree Depth for Decision Tree Surrogate Model
For KLIME interpretations, specify the depth that you want for your decision tree surrogate model. The tree depth value can be a value from 2-5 and defaults to 3. For LIME-SUP interpretations, specify the LIME-SUP tree depth. This can be a value from 2-5 and defaults to 3.
mli_vars_to_pdp¶
Number of Features for Partial Dependence Plot
Specify the maximum number of features to use when building the Partial Dependence Plot. Use -1 to calculate Partial Dependence Plot for all features. By default, this value is set to 10.
mli_nfolds¶
Cross-validation Folds for Surrogate Models
Specify the number of surrogate cross-validation folds to use (from 0 to 10). When running experiments, Driverless AI automatically splits the training data and uses the validation data to determine the performance of the model parameter tuning and feature engineering steps. For a new interpretation, Driverless AI uses 3 cross-validation folds by default for the interpretation.
mli_qbin_count¶
Number of Columns to Bin
Specify the number of columns to bin. This value defaults to 0.
mli_custom¶
Add to config.toml via TOML String
Use this input field to add to the Driverless AI server config.toml configuration file with TOML string.
mli_enable_mojo_scorer¶
Allow Use of MOJO Scoring Pipeline
Use this option to disable MOJO scoring pipeline. Scoring pipeline is chosen automatically (from MOJO and Python pipelines) by default. In case of certain models, MOJO vs. Python choice can impact pipeline performance and robustness.
mli_sample_size¶
Sample Size for Surrogate Models
When the number of rows is above this limit, sample for surrogate models. The default value is 100000.
mli_shapley_sample_size¶
Sample Size for Shapley (Original & Transformed)
When the number of rows is above this limit, sample for the MLI Shapley calculation. The default value is 100000.
mli_sequential_task_execution¶
Enable Sequential Explainers Execution (Parallel Execution When Disabled)
Specify whether to enable sequential explainers execution. This setting is enabled by default. When this setting is disabled, parallel execution is used.
mli_dia_sample_size¶
Sample Size for Disparate Impact Analysis
When the number of rows is above this limit, sample for Disparate Impact Analysis (DIA). The default value is 100000.
mli_pd_sample_size¶
Sample Size for Partial Dependence Plot
When number of rows is above this limit, sample for the Driverless AI partial dependence plot. The default value is 25000.
mli_pd_numcat_num_chart¶
Unique Feature Values Count Driven Partial Dependence Plot Binning and Chart Selection
Specify whether to use dynamic switching between PDP numeric and categorical binning and UI chart selection in cases where features were used both as numeric and categorical by the experiment. This is enabled by default.
mli_pd_numcat_threshold¶
Threshold for PD/ICE Binning and Chart Selection
If mli_pd_numcat_num_chart is enabled, and if the number of unique feature values is greater than the threshold, then numeric binning and chart is used. Otherwise, categorical binning and chart is used. The default threshold value is 11.
mli_sa_sampling_limit¶
Sample Size for Sensitivity Analysis (SA)
When the number of rows is above this limit, sample for Sensitivity Analysis (SA). The default value is 500000.
mli_nlp_sample_limit¶
Sample Size for NLP Surrogate Models
Specify the maximum number of records on which to perform MLI NLP. The default value is 10000.
klime_cluster_col¶
k-LIME Clustering Columns
For k-LIME interpretations, optionally specify which columns to have k-LIME clustering applied to.
Note: This setting is not found in the config.toml file.
qbin_cols¶
Quantile Binning Columns
For k-LIME interpretations, specify one or more columns to generate decile bins (uniform distribution) to help with MLI accuracy. Columns selected are added to top n columns for quantile binning selection. If a column is not numeric or not in the dataset (transformed features), then the column will be skipped.
Note: This setting is not found in the config.toml file.
AutoDoc Tab¶
autodoc_report_name¶
AutoDoc Name
Specify the name of the AutoDoc.
autodoc_template¶
AutoDoc Template Location
Specify the AutoDoc template path. Provide the full path to your custom AutoDoc template. To generate the standard AutoDoc, leave this field empty.
autodoc_output_type¶
AutoDoc File Output Type
Specify the AutoDoc file output type. Choose from docx (the default value) and md.
autodoc_subtemplate_type¶
AutoDoc Sub-Template Type
Specify the type of sub-templates to use. Choose from the following:
auto (Default)
md
docx
autodoc_max_cm_size¶
Confusion Matrix Max Number of Classes
Specify the maximum number of classes in the confusion matrix. This value defaults to 10.
autodoc_num_features¶
Number of Top Features to Document
Specify the number of top features to display in the document. To disable this setting, specify -1. This is set to 50 by default.
autodoc_min_relative_importance¶
Minimum Relative Feature Importance Threshold
Specify the minimum relative feature importance in order for a feature to be displayed. This value must be a float >= 0 and <= 1. This is set to 0.003 by default.
autodoc_include_permutation_feature_importance¶
Permutation Feature Importance
Specify whether to compute permutation-based feature importance. This is disabled by default.
autodoc_feature_importance_num_perm¶
Number of Permutations for Feature Importance
Specify the number of permutations to make per feature when computing feature importance. This is set to 1 by default.
autodoc_feature_importance_scorer¶
Feature Importance Scorer
Specify the name of the scorer to be used when calculating feature importance. Leave this setting unspecified to use the default scorer for the experiment.
autodoc_pd_max_rows¶
PDP and Shapley Summary Plot Max Rows
Specify the number of rows shown for the partial dependence plots (PDP) and Shapley values summary plot in the AutoDoc. Random sampling is used for datasets with more than the autodoc_pd_max_rows limit. This value defaults to 10000.
autodoc_pd_max_runtime¶
PDP Max Runtime in Seconds
Specify the maximum number of seconds Partial Dependency computation can take when generating a report. Set to -1 for no time limit.
autodoc_out_of_range¶
PDP Out of Range
Specify the number of standard deviations outside of the range of a column to include in partial dependence plots. This shows how the model reacts to data it has not seen before. This is set to 3 by default.
autodoc_num_rows¶
ICE Number of Rows
Specify the number of rows to include in PDP and ICE plots if individual rows are not specified. This is set to 0 by default.
autodoc_population_stability_index¶
Population Stability Index
Specify whether to include a population stability index if the experiment is a binary classification or regression problem. This is disabled by default.
autodoc_population_stability_index_n_quantiles¶
Population Stability Index Number of Quantiles
Specify the number of quantiles to use for the population stability index. This is set to 10 by default.
autodoc_prediction_stats¶
Prediction Statistics
Specify whether to include prediction statistics information if the experiment is a binary classification or regression problem. This value is disabled by default.
autodoc_prediction_stats_n_quantiles¶
Prediction Statistics Number of Quantiles
Specify the number of quantiles to use for prediction statistics. This is set to 20 by default.
autodoc_response_rate¶
Response Rates Plot
Specify whether to include response rates information if the experiment is a binary classification problem. This is disabled by default.
autodoc_response_rate_n_quantiles¶
Response Rates Plot Number of Quantiles
Specify the number of quantiles to use for response rates information. This is set to 10 by default.
autodoc_gini_plot¶
Show GINI Plot
Specify whether to show the GINI plot. This is disabled by default.
autodoc_enable_shapley_values¶
Enable Shapley Values
Specify whether to show Shapley values results in the AutoDoc. This is enabled by default.
autodoc_global_klime_num_features¶
Global k-LIME Number of Features
Specify the number of features to show in a k-LIME global GLM coefficients table. This value must be an integer greater than 0 or -1. To show all features, set this value to -1.
autodoc_global_klime_num_tables¶
Global k-LIME Number of Tables
Specify the number of k-LIME global GLM coefficients tables to show in the AutoDoc. Set this value to 1 to show one table with coefficients sorted by absolute value. Set this value to 2 to show two tables - one with the top positive coefficients and another with the top negative coefficients. This value is set to 1 by default.
autodoc_data_summary_col_num¶
Number of Features in Data Summary Table
Specify the number of features to be shown in the data summary table. This value must be an integer. To show all columns, specify any value lower than 1. This is set to -1 by default.
autodoc_list_all_config_settings¶
List All Config Settings
Specify whether to show all config settings. If this is disabled, only settings that have been changed are listed. All settings are listed when enabled. This is disabled by default.
autodoc_keras_summary_line_length¶
Keras Model Architecture Summary Line Length
Specify the line length of the Keras model architecture summary. This value must be either an integer greater than 0 or -1. To use the default line length, set this value to -1 (default).
autodoc_transformer_architecture_max_lines¶
NLP/Image Transformer Architecture Max Lines
Specify the maximum number of lines shown for advanced transformer architecture in the Feature section. Note that the full architecture can be found in the appendix.
autodoc_full_architecture_in_appendix¶
Appendix NLP/Image Transformer Architecture
Specify whether to show the full NLP/Image transformer architecture in the appendix. This is disabled by default.
autodoc_coef_table_appendix_results_table¶
Full GLM Coefficients Table in the Appendix
Specify whether to show the full GLM coefficient table(s) in the appendix. This is disabled by default.
autodoc_coef_table_num_models¶
GLM Coefficient Tables Number of Models
Specify the number of models for which a GLM coefficients table is shown in the AutoDoc. This value must be -1 or an integer >= 1. Set this value to -1 to show tables for all models. This is set to 1 by default.
autodoc_coef_table_num_folds¶
GLM Coefficient Tables Number of Folds Per Model
Specify the number of folds per model for which a GLM coefficients table is shown in the AutoDoc. This value must be be -1 (default) or an integer >= 1 (-1 shows all folds per model).
autodoc_coef_table_num_coef¶
GLM Coefficient Tables Number of Coefficients
Specify the number of coefficients to show within a GLM coefficients table in the AutoDoc. This is set to 50 by default. Set this value to -1 to show all coefficients.
autodoc_coef_table_num_classes¶
GLM Coefficient Tables Number of Classes
Specify the number of classes to show within a GLM coefficients table in the AutoDoc. Set this value to -1 to show all classes. This is set to 9 by default.
autodoc_num_histogram_plots¶
Number of Histograms to Show
Specify the number of top features for which to show histograms. This is set to 10 by default.