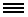The Simons Foundation Autism Research Initiative (SFARI) Investigator program supports nearly 200 researchers who are carrying out bold, innovative autism research. In 2014, these researchers published dozens of papers, encompassing genetics research, imaging studies of individuals with autism, behavioral studies and a host of other approaches to understanding the complex disorder. The following are some highlights from SFARI Investigators’ research activities in the past year.
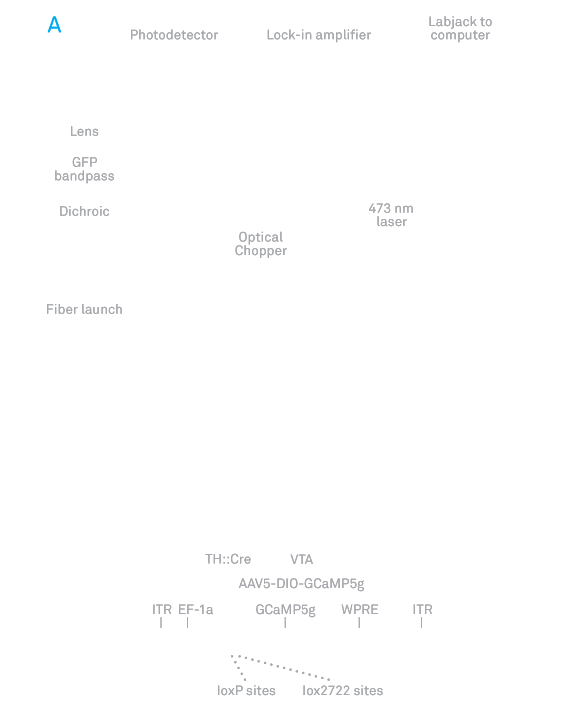
Social circuitry: Karl Deisseroth and his colleagues (A) developed a technique to measure real-time neural activity in freely moving mice and (B) found that a specific circuit that projects to the nucleus accumbens tends to show more activity (activity shown with blue dashes and colored boxes) when the mice exhibit social interactions, which are often affected in autism. Read full article
Insufficient Pruning
As children with autism transition into adolescence, their neurons prune far fewer dendritic spines — the protrusions that receive messages from other neurons — than in children without the disorder, according to a study in the September 3, 2014, issue of Neuron.
The study’s researchers, led by SFARI Investigator David Sulzer of Columbia University in New York City, examined postmortem brain samples from 16 children and teenagers with autism and 12 controls. The control teenagers had 45 percent fewer dendritic spines than their child counterparts did, whereas the teenagers with autism had only 15 percent fewer dendritic spines than the children with autism did. The adolescents with autism also had unusually high levels of mTOR, a protein that inhibits autophagy, the process by which cells recycle unneeded parts.
The team also examined a mouse model that has unusually high levels of mTOR activity and exhibits autism-like social behavior. As with the humans, the adolescent mice had a higher density of dendritic spines than did control mice, as well as a lower level of LC3-II, a marker for autophagy. When the mice were treated with rapamycin, a drug that inhibits mTOR activity, their spine density and LC3-II levels became normal, and their social behavior became similar to that of controls, suggesting that drugs that restore autophagy could be a promising approach for treating autism.

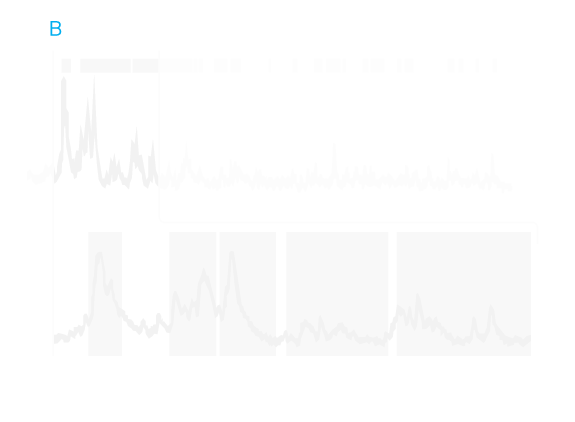
Pinpointing brain circuits: Thomas Südhof’s lab used a motor-learning task akin to log-rolling to model a cardinal feature of autism: acquired repetitive behavior. The researchers found (A) that absence of the autism-associated gene NL3 (B-G) in the dorsal striatum, a brain region known for its role in motor function, surprisingly did not change performance on the task. (H-M) Rather, deletion of NL3 in the nucleus accumbens, an area associated with learning and motivation, seemed to enhance learned repetitive behavior.
Read Full ArticleA Clear Autism Subtype
Individuals with mutations in the gene CHD8 — the autism candidate gene with by far the strongest evidence — have a consistent and recognizable phenotype, a new study shows. The study is one of the first to examine people with autism who all have the same mutation, an approach that may eventually help researchers zero in on personalized treatments for the highly heterogeneous disorder, says SFARI Investigator Evan Eichler of the University of Washington, who led the study.
The researchers collected detailed observations from 15 individuals with CHD8 mutations, 13 of whom have autism diagnoses (the other two have been diagnosed with intellectual disability and may well have undiagnosed autism, the researchers say). Most of these individuals have wide-set eyes, large ears, and broad foreheads and noses. Twelve have enlarged heads, 12 have digestive problems, especially constipation, and 10 have sleep problems. To gain insight into the biological mechanism underlying this phenotype, the researchers blocked CHD8 expression in zebrafish embryos. Like the people with CHD8 mutations, the zebrafish developed wide-set eyes and a sluggish digestive tract, and had only half as many neurons in their gut as controls did.
Attempts to identify subtypes of autism by looking at people with similar symptoms have not been very successful, the researchers wrote in the July 17, 2014 issue of Cell. The study, which indicates that CHD8 disruption is a distinct autism subtype, suggests that a ‘gene-first’ approach may be more fruitful, Eichler says.
Read Full ArticlePinpointing Brain Circuits
Two Stanford University studies have linked autism with a brain region called the nucleus accumbens, which is involved in goal-related behavior. Combined, the studies identify a particular circuit that seems to be involved in two core domains of autism, social and repetitive behaviors, giving researchers a toehold on the largely unsolved problem of understanding the neural circuitry of autism.
One study, led by SFARI Investigators Thomas Südhof and Robert Malenka, suggests that the region plays a role in repetitive behaviors in mice. The researchers looked at mice with mutations that either knock out or greatly lower the level of neuroligin-3 (NLGN3), a protein involved in synapse formation that has been strongly linked to autism. The team reported in the July 3, 2014, issue of Cell that both kinds of mutations cause mice to perform unusually well in tests of learned repetitive routines akin to log-rolling contests.
The researchers next removed NLGN3 from specific brain cells, to try to home in on the part of the brain underlying this repetitive behavior. Their study implicated a set of neurons in the nucleus accumbens that express a receptor for the chemical messenger dopamine, which is involved in reward signals. When the researchers restored NLGN3 to these neurons, the behavior of the mice returned to normal, suggesting that this type of repetitive behavior might be amenable to drug therapies.
In the second study, researchers developed a new technique that provides scientists with a window into the dynamics of real-time circuit activity during social interactions in mice, and points to a specific circuit whose activity encodes such interactions. The technique, created by a team led by SFARI Investigator Karl Deisseroth and Robert Malenka, involves inserting a tiny fiber-optic probe into the brains of mice that have been engineered to express a fluorescent molecule during the firing of neurons that make dopamine. When the probe was placed in the nucleus accumbens, it measured that significantly more dopamine spikes occurred when the mice interacted with new mice than when they interacted with novel objects, the researchers reported June 19, 2014, in Cell. The results suggest that a circuit connecting a region called the ventral tegmental area, a major source of dopamine neurons, to the nucleus accumbens specifically modulates social behavior.
Read Full ArticleRegulating Brain Size
A chromosomal region called 16p11.2, which is strongly linked to autism, may control brain size, researchers reported August 20, 2014, in The Journal of Neuroscience.
Deletions and duplications in 16p11.2, which contains 29 genes, are among the genetic variations that have been most frequently associated with autism. To understand the implications of these deletions and duplications, in 2010 the Simons Foundation created the Simons Variation in Individuals Project (Simons VIP, see Simons Simplex Collection), which, like Eichler’s study of individuals with CHD8 mutations, takes a ‘gene-first’ approach by studying the phenotypes of several hundred individuals with 16p11.2 deletions or duplications and their families.
In the August study, a research team led by SFARI Investigator Randy Buckner of Harvard University used magnetic resonance imaging to scan the brains of a cohort of individuals in the Simons VIP collection, along with a control group. Deletions in 16p11.2 increased brain size by about 9 percent, the researchers found, while duplications reduced brain size by a similar amount. Changes in brain size were observed across all brain regions, and within each region the deletion and the duplication produced comparable changes in opposite directions, suggesting that the effect of 16p11.2 on brain size is dose dependent. Within the cortex, surface area, which is determined early in development, was affected more strongly than thickness, which is determined later on, suggesting that the mechanisms underlying the mutations’ effects may come into play in early embryonic development.
Read Full ArticleIncorrect Splicing
Depending on which cell type it is in, the same gene can give rise to many different proteins, as different portions of the gene’s sequence get spliced out while the gene is being transcribed to RNA. One of the RNA-binding proteins that regulates this splicing in the nervous system, RBFOX, has been implicated in autism, but until now it has been difficult to identify just which genes RBFOX regulates.
In a study published March 27, 2014 in Cell Reports, a team led by SFARI Investigators Robert Darnell of Rockefeller University and Chaolin Zhang of Columbia University made a complete map, at single-nucleotide resolution, of all RBFOX’s RNA interaction sites in the mouse brain. Of the 1,059 RBFOX splicing events the team identified, more than 10 percent involve autism risk genes, a far greater proportion than chance would predict. The work suggests that RBFOX might be a ‘hub’ that regulates many autism risk genes.
RBFOX is actually a family of three proteins, the products of a single gene, that play similar roles in the regulation of mRNA splicing: In postmortem brain tissue of some individuals with autism, the researchers found that all three RBFOX proteins were present at lower-than-normal levels, suggesting that mRNA splicing critical for normal brain function was altered in these individuals.
Read Full Article