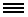To Olga Troyanskaya, leader of the genomics group at SCDA, figuring out how to use big data to study complex disorders such as autism or cancer is like trying to develop a Google for genomics. “Before Google and other ‘smart’ search engines, the Internet was a collection of directories with no clear assessment of quality or relevance, and written in different languages,” she says. “Genomic data are even more complicated: They represent hundreds of diseases, tissues and clinical treatments, and are made by more than 50 different technologies. How can one identify and integrate relevant information across all these datasets?”

Brain-specific functional gene networks can illuminate the molecular basis of neurodevelopmental disorders. Above, a section of a brain network thought to be relevant to autism.
Troyanskaya’s team develops algorithms that can spot similar patterns in gene expression across many different kinds of tissue and disease, regardless of the technology used to gather the data. For example, the same genetic pathways that are important in neurons in the brain also exist in kidney cells, so kidney disease data might actually teach us something about brain disorders such as autism. “It’s very counterintuitive,” she admits. “It’s not based on symptoms or single-gene mutations. It’s only algorithmically that you can systematically identify such signals.”
This “messy gold mine” of gene expression data, as Troyanskaya calls it, could unlock new understanding of complex disorders by uncovering genetic links that were previously invisible. Any biological experiment inevitably perturbs many different aspects of a cell’s function, and the Troyanskaya group’s methods put those inevitable extra perturbations to good scientific use by first identifying patterns in these ‘noisy’ datasets that are useful outside of the original experimental context in which they occurred, and then aggregating these datasets together.
According to Troyanskaya, autism research lends itself especially well to this approach precisely because there is no ‘autism gene’ to pinpoint in isolation. Instead, autism is a networked disorder whose symptoms are associated with the coordinated behavior of multiple genes. While damage to a single gene can have major impact, this impact is most likely modified by small differences between individuals in expression and function of other genes in the network. Troyanskaya’s computational analysis allows every human gene to be ranked based on how likely it is to be associated with autism based on its functional role in the brain’s molecular networks.
As her team uncovers these associations in collaboration with SFARI, they also build software that lets other researchers apply the same algorithmic methods to explore other open questions in cell biology and medicine. “We’re working across diverse tissues and cell types — looking at large collections of biological data, and figuring out algorithms that are able to isolate the relevant signals in a very accurate way,” she says. “The philosophy of my group is that with smart algorithms, more data is always better.”