class: title-slide background-size: cover <h1 class="center middle">Raster Data and the {raster} package</h1> <h3 class="center middle"></h3> <div class="logo"></div> <div class="databird"></div> --- # The `raster` package is 10 years old!  --- # But it has been around for so long for a reason * It's powerful * It's intuitive (at least I think so) * Robert keeps it up to date --- # Robert J. Hijmans  --- # Fully updated intro vignette [This is a useful reference.](https://cran.r-project.org/web/packages/raster/vignettes/Raster.pdf) --- # As discussed earlier, "raster" refers to gridded data --- # And raster data can be single-band or multiple-band -- 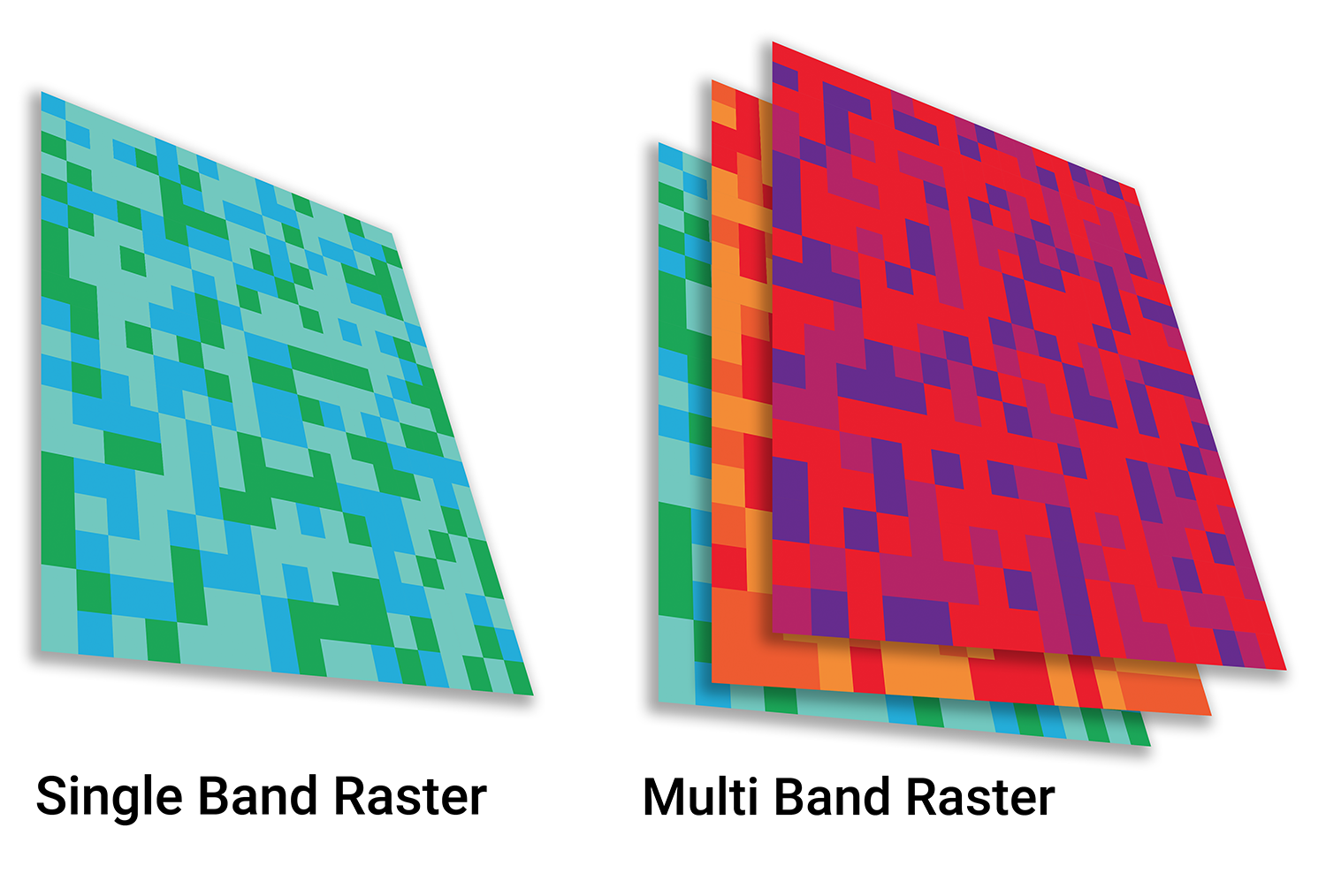 --- # Read a single-band raster with `raster()` -- ```r elevation <- raster("data/elevation.tif", quiet = TRUE) ``` --- # Single-band class is `RasterLayer` -- ```r class(elevation) ``` ``` ## [1] "RasterLayer" ## attr(,"package") ## [1] "raster" ``` --- # Map the elevation data -- ```r tm_shape(elevation) + tm_raster() ``` -- 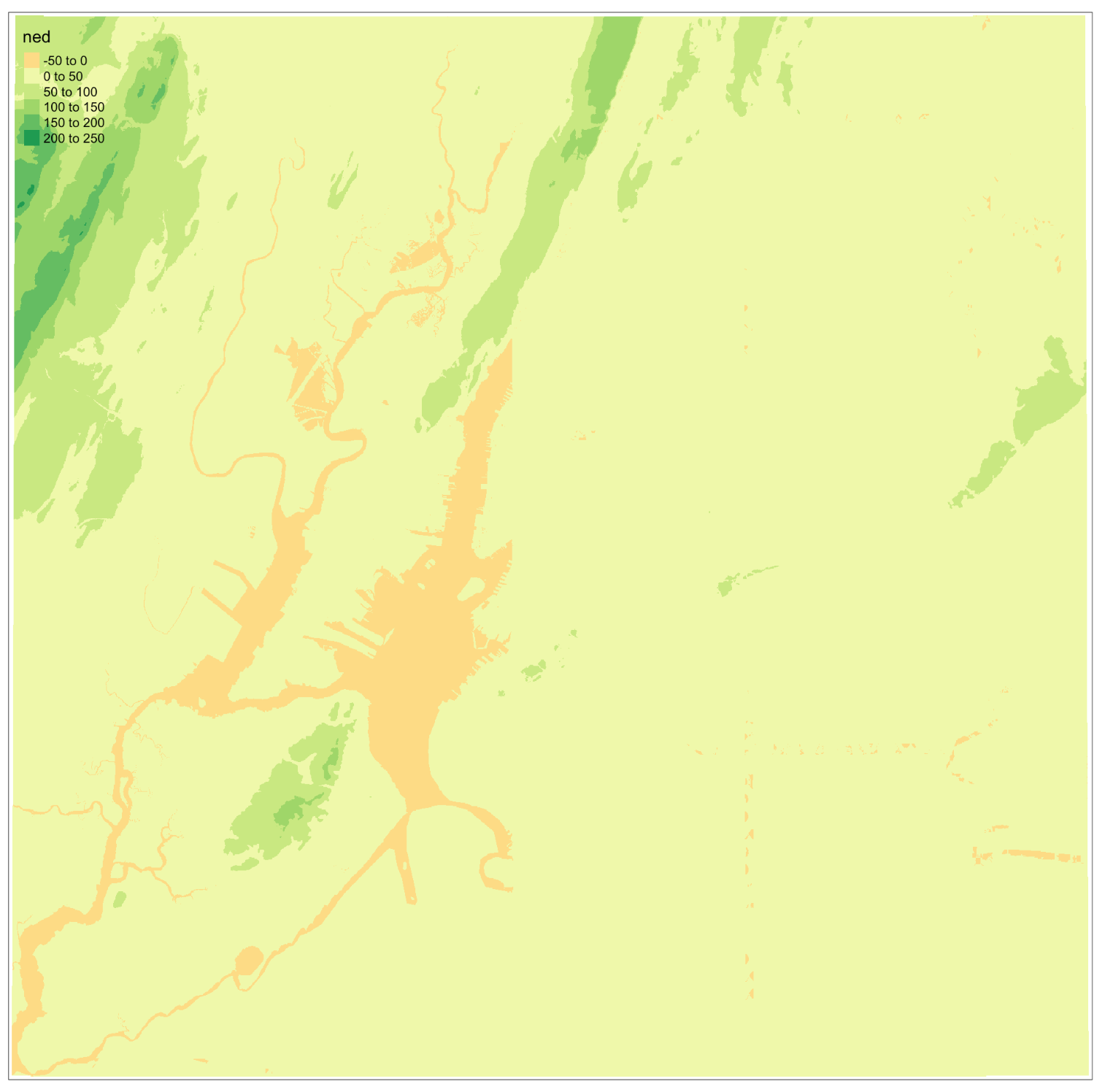 --- # Our `RasterLayer` metadata -- ```r elevation ``` ``` ## class : RasterLayer ## dimensions : 1530, 2039, 3119670 (nrow, ncol, ncell) ## resolution : 77, 101 (x, y) ## extent : 911860, 1068863, 119161.4, 273691.4 (xmin, xmax, ymin, ymax) ## crs : +proj=lcc +lat_1=41.03333333333333 +lat_2=40.66666666666666 +lat_0=40.16666666666666 +lon_0=-74 +x_0=300000.0000000001 +y_0=0 +ellps=GRS80 +towgs84=0,0,0,0,0,0,0 +units=us-ft +no_defs ## source : /Volumes/GoogleDrive/My Drive/projects/workshop-r-spatial-slides/big-project-files/gis-data/nyc/processed/ned.tif ## names : ned ## values : -14.05926, 209.7131 (min, max) ``` --- # Read a multi-band raster with `brick()` -- ```r satellite_image <- brick("data/satellite.tif", quiet = TRUE) ``` --- # Multi-band class is `RasterBrick` -- ```r class(satellite_image) ``` ``` ## [1] "RasterBrick" ## attr(,"package") ## [1] "raster" ``` --- # Map multi-band raster with `tm_raster()` -- This is an image raster with a red, green and blue layer. What happens here? ```r tm_shape(satellite_image) + tm_raster() ``` -- 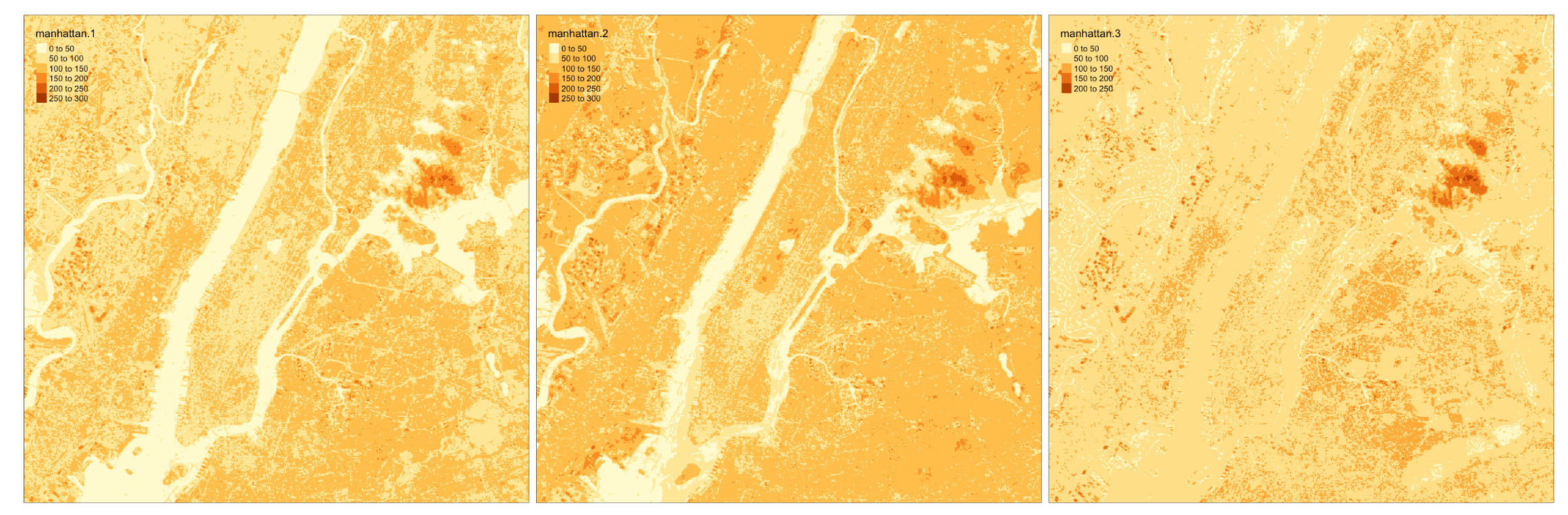 --- # Map our multi-band image with `tm_rgb()` -- ```r tm_shape(satellite_image) + tm_rgb() ``` <img src="6-raster-data_files/figure-html/unnamed-chunk-12-1.png" style="display: block; margin: auto;" /> --- # The metadata in a `RasterBrick` object is very similar -- ```r satellite_image ``` ``` ## class : RasterBrick ## dimensions : 773, 801, 619173, 3 (nrow, ncol, ncell, nlayers) ## resolution : 29.98979, 30.00062 (x, y) ## extent : 575667.9, 599689.7, 4503277, 4526468 (xmin, xmax, ymin, ymax) ## crs : +proj=utm +zone=18 +datum=WGS84 +units=m +no_defs +ellps=WGS84 +towgs84=0,0,0 ## source : /Users/zevross/git-repos/workshop-r-spatial-slides/data/nyc/manhattan.tif ## names : manhattan.1, manhattan.2, manhattan.3 ## min values : 0, 0, 0 ## max values : 255, 255, 255 ``` --- # Get the names of your layers with `names()` ```r names(satellite_image) ``` ``` ## [1] "manhattan.1" "manhattan.2" "manhattan.3" ``` --- # Two options for extracting a layer of your brick -- ```r satellite_image$manhattan.1 ``` -- ```r subset(satellite_image, "manhattan.1") ``` --- exclude:true # RasterStack ??? The principal difference between these two classes is that a RasterBrick can only be linked to a single (multi-layer) file. In contrast, a RasterStack can be formed from separate files and/or from a few layers (‘bands’) from a single file.In fact, a RasterStack is a collection of RasterLayer objects with the same spatial extent and resolution. In essence it is a list of RasterLayer objects. A RasterStack can easily be formed form a collection of files in different locations and these can be mixed with RasterLayer objects that only exist in the RAM memory (not on disk).A RasterBrick is truly a multi-layered object, and processing a RasterBrick can be more efficient than processing a RasterStack representing the same data. However, it can only refer to a single file. A typical example of such a file would be a multi-band satellite image or the output of a global climate model (with e.g., a time series of temperature values for each day of the year for each raster cell). Methods that operate on RasterStack and RasterBrick objects typically return a RasterBrick object. --- class: center, middle # You can also use `raster()` and `brick()` to create a raster from scratch --- # Create a boring raster -- ```r r <- raster(nrow = 10, ncol = 10) ``` --- # Try to plot your boring raster -- ```r plot(r) ``` ``` ## Error in .plotraster2(x, col = col, maxpixels = maxpixels, add = add, : no values associated with this RasterLayer ``` --- # Why no plot? --- # Use the `getValues()` function to look at the values -- ```r getValues(r) ``` ``` ## [1] NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA ## [24] NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA ## [47] NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA ## [70] NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA NA ## [93] NA NA NA NA NA NA NA NA ``` --- # Can't plot a raster with no values --- # Use `values()` to add values -- ```r values(r) <- rnorm(100) # assign random normal data ``` -- ```r # You can also include values when creating r <- raster(nrow = 10, ncol = 10, vals = rnorm(100)) ``` --- # Now we can plot -- ```r plot(r, axes = FALSE) ``` <img src="6-raster-data_files/figure-html/unnamed-chunk-22-1.png" style="display: block; margin: auto;" /> --- # Note that your cell values cannot be characters -- ```r my_letters <- sample(LETTERS, 100, replace = TRUE) head(my_letters) ``` ``` ## [1] "R" "Q" "C" "X" "D" "K" ``` -- ```r values(r) <- my_letters ``` ``` ## Error in setValues(x, value): values must be numeric, integer, logical or factor ``` --- # But they can be factors -- ```r values(r) <- factor(my_letters) ``` -- ```r tm_shape(r) + tm_raster() ```  --- # Technically the values stored are integers mapped to a table of levels -- ```r getValues(r)[1:10] ``` ``` ## [1] 18 17 3 24 4 11 1 5 22 19 ``` --- # The levels of the categorical raster can be accessed with `levels()` -- ```r levels(r)[[1]][1:5,] ``` ``` ## ID VALUE ## 1 1 A ## 2 2 B ## 3 3 C ## 4 4 D ## 5 5 E ``` --- # A brick can be created by stacking `RasterLayers` with `brick()` --- # Perhaps the most boring brick ever created... ```r b <- brick(r, r, r) b ``` ``` ## class : RasterBrick ## dimensions : 10, 10, 100, 3 (nrow, ncol, ncell, nlayers) ## resolution : 36, 18 (x, y) ## extent : -180, 180, -90, 90 (xmin, xmax, ymin, ymax) ## crs : +proj=longlat +datum=WGS84 +ellps=WGS84 +towgs84=0,0,0 ## source : memory ## names : layer.1, layer.2, layer.3 ## min values : 1, 1, 1 ## max values : 26, 26, 26 ``` --- # Where, on earth, are those rasters we just created? --- # By default, those hand-created rasters span the globe And have an unprojected CRS of WGS84 --- # To prove this, read in the countries with {rnaturalearth} for context -- ```r countries <- rnaturalearth::ne_countries() ``` --- # Map our random letters raster with countries on top -- ```r tm_shape(r) + tm_raster(alpha = 0.5) + tm_shape(countries) + tm_borders(lwd=2) ``` 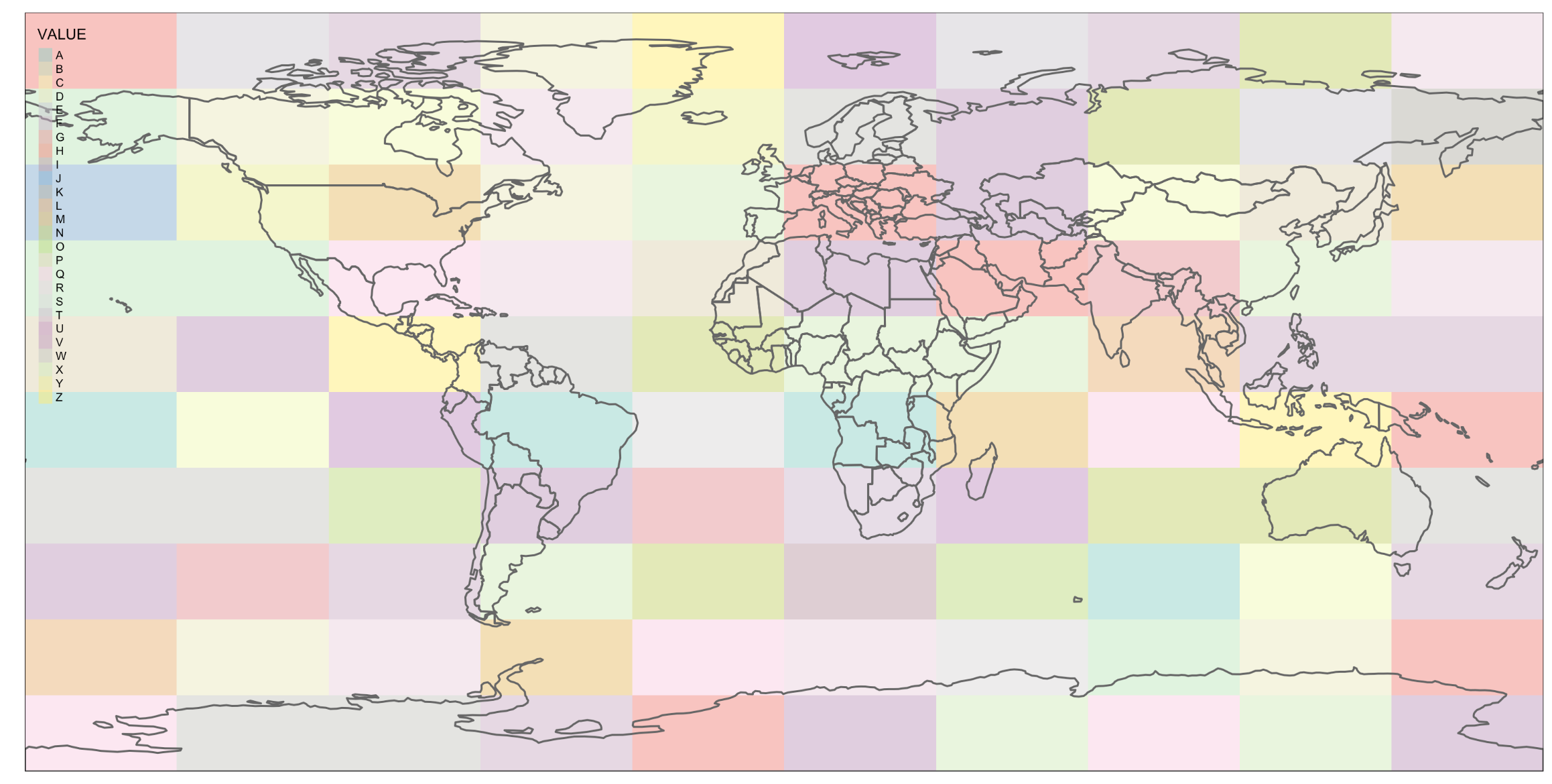 --- # But if you want your raster in a specific place on earth --- # You can use another object to define the extent -- ```r boroughs <- read_sf("boroughs.gpkg") ``` --- # Supply the object to `raster()` -- ```r rand <- raster(boroughs, ncols = 10, nrows = 10, vals = rnorm(100)) ``` --- # Plot our random raster ```r tm_shape(rand) + tm_raster(n = 100, legend.show = FALSE) + tm_shape(boroughs) + tm_borders(lwd = 3, col = "black") ``` <img src="6-raster-data_files/figure-html/unnamed-chunk-34-1.png" width="65%" style="display: block; margin: auto;" /> --- class: center, middle # Extracting information about your rasters --- # The console printout shows the metadata ```r elevation ``` ``` ## class : RasterLayer ## dimensions : 1530, 2039, 3119670 (nrow, ncol, ncell) ## resolution : 77, 101 (x, y) ## extent : 911860, 1068863, 119161.4, 273691.4 (xmin, xmax, ymin, ymax) ## crs : +proj=lcc +lat_1=41.03333333333333 +lat_2=40.66666666666666 +lat_0=40.16666666666666 +lon_0=-74 +x_0=300000.0000000001 +y_0=0 +ellps=GRS80 +towgs84=0,0,0,0,0,0,0 +units=us-ft +no_defs ## source : /Volumes/GoogleDrive/My Drive/projects/workshop-r-spatial-slides/big-project-files/gis-data/nyc/processed/ned.tif ## names : ned ## values : -14.05926, 209.7131 (min, max) ``` --- # You can access the "slots" (attributes) directly -- ```r elevation@crs ``` ``` ## CRS arguments: ## +proj=lcc +lat_1=41.03333333333333 +lat_2=40.66666666666666 ## +lat_0=40.16666666666666 +lon_0=-74 +x_0=300000.0000000001 +y_0=0 ## +ellps=GRS80 +towgs84=0,0,0,0,0,0,0 +units=us-ft +no_defs ``` -- ```r elevation@data@max ``` ``` ## [1] 209.7131 ``` --- # But there are utility functions to make it easier -- ```r crs(elevation) ``` ``` ## CRS arguments: ## +proj=lcc +lat_1=41.03333333333333 +lat_2=40.66666666666666 ## +lat_0=40.16666666666666 +lon_0=-74 +x_0=300000.0000000001 +y_0=0 ## +ellps=GRS80 +towgs84=0,0,0,0,0,0,0 +units=us-ft +no_defs ``` -- ```r maxValue(elevation) ``` ``` ## [1] 209.7131 ``` --- # Many more useful utility functions * `extent()` * `ncell()` * `nlayers()` * `filename()` * `crs()` * `minValue()` --- # Get cell value counts with `freq()` -- ```r r <- raster(nrow = 100, ncol = 100, vals = sample(1:10, 100*100, replace = TRUE)) ``` -- ```r freq(r) %>% head() ``` ``` ## value count ## [1,] 1 1071 ## [2,] 2 980 ## [3,] 3 981 ## [4,] 4 1006 ## [5,] 5 981 ## [6,] 6 986 ``` --- class: center, middle # {raster} can work with large rasters --- # The elevation raster has a lot of grid cells ```r ncell(elevation) ``` ``` ## [1] 3119670 ``` --- # More than 3 million cells but in R it takes little memory ```r pryr::object_size(elevation) ``` ``` ## 11.6 kB ``` --- # On disk, the file is not small ```r # Size on my computer fs::file_info("elevation.tif")$size ``` ``` ## 10.2M ``` --- # Raster values are not read by default * Rasters can be very big * To conserve memory raster values are imported only when required * For large rasters, data is processed in chunks --- ## The `inMemory()` function tells you if the raster values have been read into R ```r inMemory(satellite_image) ``` ``` ## [1] FALSE ``` --- ## Read values with the `getValues()` function -- ```r vals <- getValues(elevation) hist(vals) ``` <img src="6-raster-data_files/figure-html/unnamed-chunk-47-1.svg" style="display: block; margin: auto;" /> --- class: middle, center # Converting between vectors and rasters --- # Note that while there is some support for {sf} in {raster} it is not complete --- # You'll need to review the function documentation to determine what type of input is accepted --- # If a function only accepts a `Spatial*` object you can convert with `as()` -- ```r class(boroughs) ``` ``` ## [1] "sf" "tbl_df" "tbl" "data.frame" ``` -- ```r as(boroughs, "Spatial") %>% class() ``` ``` ## [1] "SpatialPolygonsDataFrame" ## attr(,"package") ## [1] "sp" ``` --- # Use the `rasterize()` function to convert vectors to raster .footnote[For large rasters this can be computationally intensive and the {fasterize} package can help speed things up] --- # You might do this for landscape analysis or raster math  .footnote[[Image source](https://www.appgeo.com/carpegeo/map_overlays/)] --- # With `rasterize()` you assign the attributes from a vector layer to a raster --- # For this example we will create a raster layer of the boroughs --- # Our boroughs vector data ```r tm_shape(boroughs) + tm_polygons("BoroName") ``` <img src="6-raster-data_files/figure-html/unnamed-chunk-50-1.svg" style="display: block; margin: auto;" /> --- # Our boroughs attribute table ```r glimpse(boroughs) ``` ``` ## Observations: 5 ## Variables: 7 ## $ BoroCode <int> 1, 2, 5, 3, 4 ## $ BoroName <chr> "Manhattan", "The Bronx", "Staten… ## $ Shape_Leng <dbl> 339736.6, 397460.6, 318700.5, 576… ## $ Shape_Area <dbl> 635147797, 1182399343, 1630762350… ## $ diso <int> 1, 1, 1, 1, 1 ## $ AreaSqMile <dbl> 22.78279, 42.41274, 58.49555, 71.… ## $ geom <MULTIPOLYGON [US_survey_foot]> MULTIPO… ``` --- # Step 1: Convert to an {sp} object After reviewing the help for `rasterize()` -- ```r boroughs_sp <- as(boroughs, "Spatial") ``` --- # Step 2: create the raster that values will be transferred to -- ```r n <- 1000 r <- raster(boroughs_sp, ncols = n, nrows = n) ``` --- # Run `rasterize()` -- ```r boro_raster <- rasterize(boroughs_sp, r, field = "BoroCode") ``` --- # Our rasterized boroughs ```r tm_shape(boro_raster) + tm_raster() + tm_layout(legend.show = FALSE) ``` <img src="6-raster-data_files/figure-html/unnamed-chunk-55-1.png" style="display: block; margin: auto;" /> --- # For converting from a raster to vectors you can use * `rasterToPolygons()` * `rasterToPoints()` --- # Create a random raster for this example -- ```r set.seed(100) r <- raster(nrow = 30, ncol = 30, vals = sample(1:3, 900, replace = TRUE)) ``` -- ```r tm_shape(r) + tm_raster() + tm_layout(legend.show = FALSE) ```  --- # Extract points from the raster As a Spatial* object with `spatial = TRUE` ```r pts <- rasterToPoints(r, spatial = TRUE) ``` -- ```r class(pts) ``` ``` ## [1] "SpatialPointsDataFrame" ## attr(,"package") ## [1] "sp" ``` --- # Convert to an {sf} object -- ```r pts <- as(pts, "sf") ``` -- ```r # This also works pts <- st_as_sf(pts) ``` --- # Plot the points -- ```r my_layout <- tm_layout(frame = FALSE, legend.show = FALSE) m1 <- tm_shape(r) + tm_raster() + my_layout m2 <- tm_shape(pts) + tm_dots("layer", size = 0.5, shape = 21) + my_layout tmap_arrange(m1, m2, nrow = 1) ``` -- 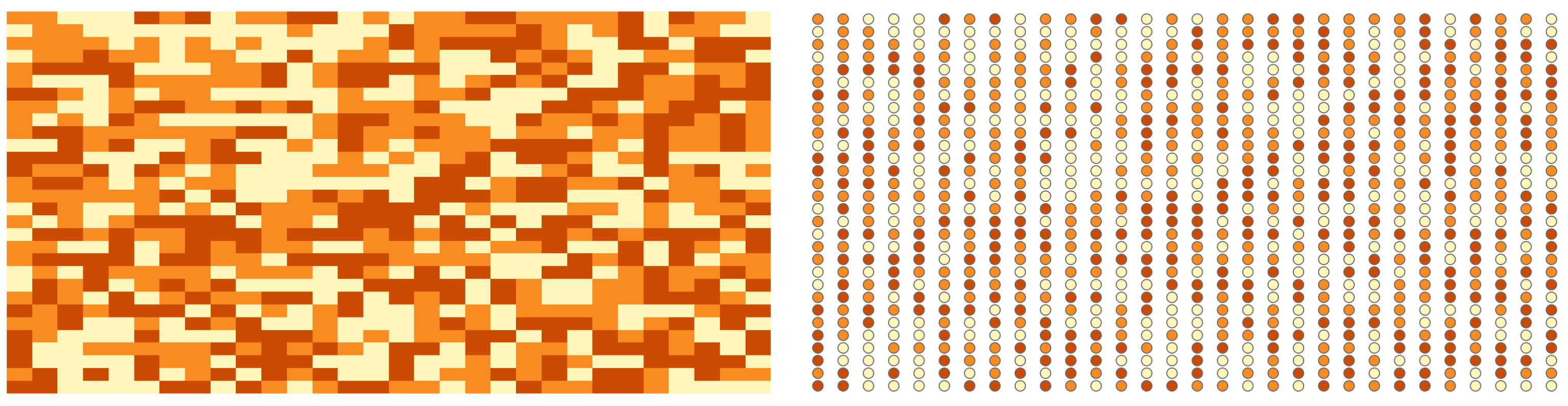 --- # Extract the polygons ```r polys <- rasterToPolygons(r, dissolve = TRUE) %>% st_as_sf(sf) ``` --- # Plot the polys ```r m3 <- tm_shape(polys) + tm_polygons("layer", border.col = "black") + my_layout tmap_arrange(m1, m3, nrow = 1) ``` 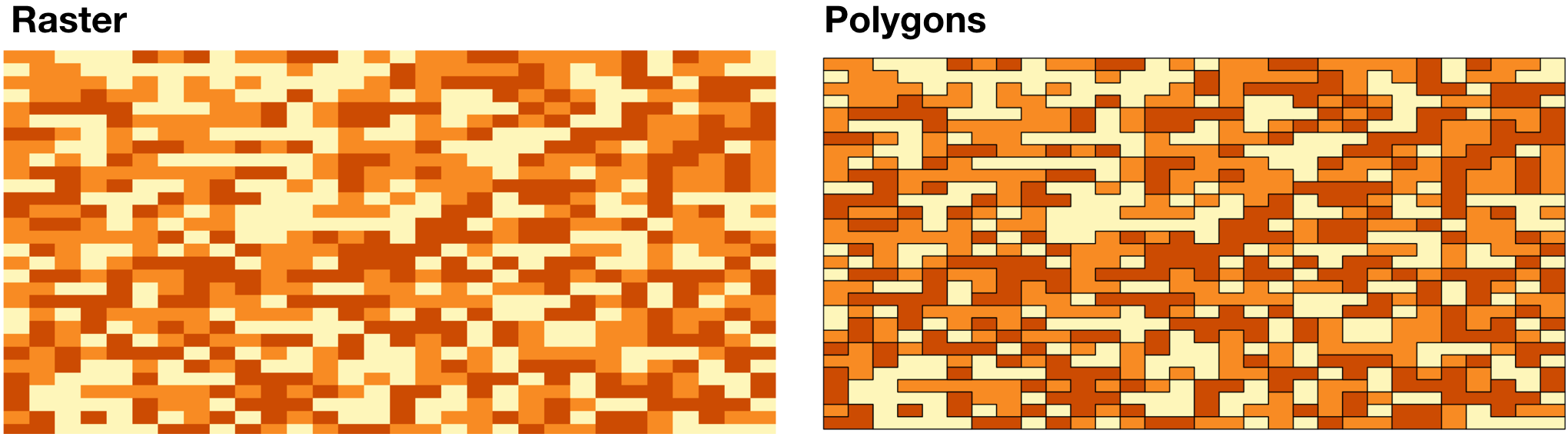 --- class: center, middle, doExercise # open_exercise(6)